In this article, we shall study the structure of DNA.
Each cell in a particular living organism contains the exact same DNA. The size of the DNA polymer is directly related to the complexity of the organism; more complex organisms tend to have larger molecules of DNA, while less complex organisms have smaller DNA. For e.g. The DNA in simple bacteria contains about 8 million nucleotides, whereas human DNA contains up to 500 million nucleotides.
Primary Structure:
The primary structure of DNA is simply the sequence of nucleotides. The sugar-phosphate chain is called the DNA backbone, and it is constant throughout the entire DNA molecule. The variable portion of DNA is the sequence of nitrogenous bases.
The phosphate groups link the 3rd carbon of one sugar (of deoxyribose or ribose) to the 5th carbon of the next sugar (of deoxyribose or ribose).
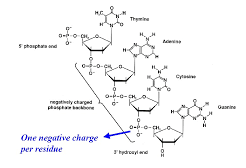
A strand of DNA has two distinct terminals or ends, one will be a 5-phosphate end and the other will be a 3-hydroxyl end. By convention, a nucleic acid sequence is always read in the 5 to 3 direction, that is, from the sugar with the free 5-phosphate to the sugar with a 3-hydroxyl group. The order of nucleotides is generally written using the capitalized first letter of the name of a base. Thus the following structure is written ACGT (in the 5 to 3 direction)
Secondary Structure (Double Helix Structure):
Double Helix Structure:
The secondary structure of DNA was proposed by James Watson and Francis Crick in 1953. This was considered to be the greatest discovery of modern biology and hence they got Nobel prize for the same. They proposed that DNA is a double helix containing two polynucleotide strands wound as if around a central axis.
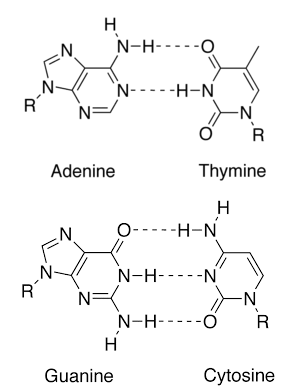
Complementary Base Pairing:
The two polynucleotide strands are connected by hydrogen bonds formed between a purine on one strand and a pyrimidine on the other. In DNA, adenine is always paired with thymine and guanine is always paired with cytosine. The pairs A-T and G-C are called complementary base pairs. According to base-pairing rules discovered by Watson and Crick, each A is bound to T and each G is bound to C.
Purine Pyrimidine Ratio:
Therefore, the total number of A’s in any molecule of DNA must be equal to a total number of T’s (the same is true of G and C). Thus, the % of A in DNA must equal the % of T (the same is true of G and C). The total percent of A, T, G, and C must be equal to 100. Human DNA contains 30% adenine, 30% thymine, 20% cytosine, and 20% guanine.
Due to complementary base pairing, the total number of purine bases is equal to a total number of pyrimidine bases. Thus the ratio is 1:1. This relation is called Chargaff’s rule.
The Polarity of Strands (Antiparallel):
The two strands of DNA run antiparallel to one another, that is, the two strands run in opposite directions. One in the 5 to 3 direction, the other in the 3 to 5 direction. Therefore, both ends of the double helix contain the 5 end of one strand (5 phosphate) and the 3 end of the other (3 OH).

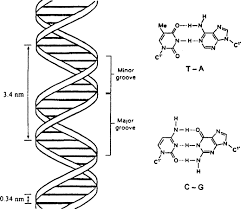
Major and Minor Grooves:
The helix is right-handed and it results in the formation of a deep groove called a major groove and another shallow groove called a minor groove. The two groves are alternate.

Dimensions:
The diameter of the DNA molecule is 2 nm. The deflection pitch angle of a helix is 36º. The helix completes its one turn after covering a distance of 3.4 nm. There are 10 base pairs in one complete spiral.
Packaging of DNA:
The length of a DNA double helix molecule in a typical mammalian cell is approximately 2.2 metres. This length can be obtained by multiplying the total number of base pairs present in the DNA double helix, which is 6.6 X 109, with the distance between two consecutive base pairs, which is 0.34 × 10-9 m. Such a long polymer is packaged within a typical nucleus of size 10 -6 m, by condensing it by coiling and supercoiling (coiled-coil) to fit in the nucleus. which is different for both prokaryotes and eukaryotes.
In prokaryotes, the negatively charged DNA is arranged in large loops and is held together by a few positively-charged proteins, called the nucleoid. In eukaryotes, a single molecule of negatively charged DNA is packaged around a pool of positively charged proteins called histones.
Histones are proteins that are rich in basic amino acid residues lysines and arginines. They carry a positive charge on their outer side chain. There are four types of histones H2A, H2B, H3 and H4. Two types of each, thus four molecules of histones form an octamer. The negatively charged DNA is wrapped around the positively charged histone octamer to form a nucleosome. It is held in place by the H1 histone.
A typical nucleosome has around two hundred (146 + 54) base pairs of DNA helix and it is the nucleosomes that make up the repeating unit in chromatin.

Under an electron microscope, the nucleus shows a chromatin network. In the network, nucleosome can be seen as beads on the string. 1 and 3/4th turn around histone octamer consist of 146 base pairs and is called core DNA. Adjacent DNA acts as a linking strand consists of 54 base pairs and is called linker DNA. H1 histone is present in the linker region and as DNA makes two complete turns it is present where DNA starts wrapping the histone and leaves it.

The thin and long nucleosome fibre is coiled again to form a supercoiled structure to make solenoid fibre with diameter 30nm or 300 A°. Nucleosome and solenoid fibres are characteristic of the nucleus at interphase. For further packing of chromatin additional protein Non-Histone Chromosomal (NHC) proteins are responsible. The chromatin fibres are of two types, euchromatin and heterochromatin.
The euchromatin fibres are of thirty to eighty nanometres in diameter and are loosely packed and stain light. While heterochromatin fibres are of about three hundred nanometres in diameter and are more densely packed and stain dark. The euchromatin is transcriptionally active, while heterochromatin is inactive.
These chromatin fibres coil further and condense to form short and thick bodies called chromosomes, which are further packaged within the nucleus.