There are three types of non-genetic RNA. They are mRNA, rRNA, and tRNA. These RNAs are produced on the DNA template by the process called transcription.
mRNA or Messenger RNA:
The mRNA molecule is always simple and straight without any fold and is transcribed from one strand of DNA. It constitutes 3 to 5 % of total RNA. It carries the base sequence complementary to the DNA template strand.
A base sequence of three nucleotides on mRNA is called a codon. Ribosomes translate these triplet codons into the amino acid sequence of the polypeptide chain to form proteins. This is called mRNA language or genetic code or cryptograms because they carry coded information for the synthesis of a particular protein.
It has two ends 5′ end and 3′ end. The codon present at 5′ end is called start codon or initiation codon. It is usually AUG, in some cases, it may be GUG. The codon at 3′ end is called stop codon or terminator codon or nonsense codon. It is either UAA, UAG or UGA. Each codon specifies a specific amino acid. End codon does not specify any amino acid, hence they are referred as nonsense codons.
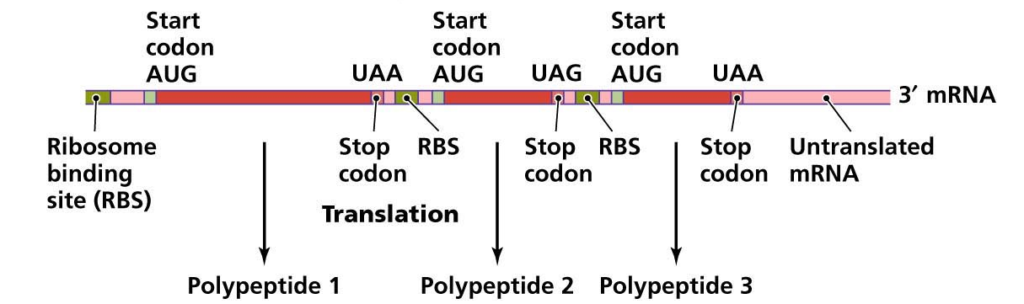
Length of mRNA:
The length of mRNA depends upon the length of the polypeptide chain it Codes for. Polypeptide length varies from a chain of a few amino acids to thousands of amino acids. The message is read in the groups of three consecutive bases from a fixed starting point up to a fixed stopping point.
Life Span of mRNA:
In bacteria, mRNA is transcribed and translated in a single cellular compartment and the two processes are so closely linked that they occur simultaneously.
Transcription begins when the enzyme RNA polymerase binds to DNA and then moves along making a copy of one strand. As soon as the transcription begins, the ribosomes attach to the 5′ end (free end) of the mRNA and start translation while the other end of mRNA is still under synthesis. This is known as coupled transcription and translation in prokaryotes.
After the translation of the whole of mRNA is completed, the mRNA is then degraded in 5′ → 3′ direction. The mRNA is synthesized, translated and degraded all in rapid succession and all in 5′ → 3′ direction. An individual mRNA molecule survives only for a minute or less.
In eukaryotes, transcription occurs in the nucleus while translation takes place in the cytoplasm. Eukaryotic mRNA is quite stable and survives from a few minutes to more than a day. In mammalian RBC, through the nucleus is lost, mRNA continues to produce haemoglobin for many days.
Coding and Non-coding Regions:
All mRNAs have two types of regions. The coding region consists of a series of codons. But the mRNA is longer than the coding regions. The length of newly synthesized mRNA is much larger than the length of mRNA used for translation. The coding regions are called exons. Between the coding regions lie various non-coding regions called introns. Genes with these intervening sequences are called Split genes or Interrupted genes.
rRNA or Ribosomal RNA:
Most of the RNA of the cell is in the form of ribosomal RNA which constitutes about 80% of the total RNA. Ribosomes consist of many types of rRNA. It is a single-stranded structure which is variously folded upon itself. In the folded region, it may show a pairing between complementary bases. It remains associated with ribosomes permanently. It provides a proper binding site for m-RNA by orienting the mRNA molecule so that all codons can be read perfectly.

The rRNA molecules form the secondary structure of double-stranded stems and single-stranded loops by extensive complementary base pairings. The rRNA plays a major role in protein synthesis. They interact with mRNA and tRNA at each step of translation or protein synthesis.
The 3′ terminus of rRNA of 16S rRNA interacts with the initiation site on mRNA which is called Shine-Dalgarno sequence and lies just before the start codon AUG.
The 23S rRNA plays an active role in peptidyl transferase activity. Movement of tRNA between A and P site on the ribosome is aided by 23 S rRNA.
The rRNA molecules form complexes with specific proteins in ribosomes. The RNA- protein complexes are called ribonucleoproteins (RNP).
tRNA or Transfer RNA:
The tRNA molecule is single-stranded and constitutes 10 to 20 % of total RNA. It is the smallest of all the RNA types. t-RNA is also called s-RNA soluble RNA because it can not be easily separated even by ultracentrifugation techniques. It delivers amino acids to the ribosome and decodes the information of mRNA. Each nucleotide triplet codon on mRNA represents an amino acid. The tRNA plays the role of an adaptor and matches each codon to its particular amino acid in the cytoplasmic pool. The structure of t-RNA is explained by two models
Clover Leaf Structure of tRNA:

The tRNA due to its property of having stretches of complementary base pairs forms the secondary structure, which is in the form of a cloverleaf.
Several regions of the single-stranded molecule form double-stranded stems or arms and single-stranded loops due to the folding of various regions of the molecule. These double-stranded stems have complementary base pairs. A typical tRNA has bases numbering from 1-76, using the standard numbering convention where position 1 is the 5′ end and 76 is the 3′ end.
- Amino acid arm: It has a seven base pairs stem formed by base pairing between 5′ and 3′ ends of tRNA. At 3′ end a sequence of 5′-CCA-3′ is added. This is called CCA arm or amino acid acceptor arm. Amino acid binds to this arm during protein synthesis.
- D-arm:
- Going from 5′ to 3′ direction or anticlockwise direction, the next arm is D-arm. It has a 3 to 4 base pair stem and a loop called D-loop or DHU-loop. It contains a modified base dihydrouracil.
- Anticodon arm: Next Topic is the arm which lies opposite to the acceptor arm. It has a five base pair stem and a loop in which there are three adjacent nucleotides called anticodon which is complementary to the codon of mRNA.
- An extra arm: Next Topic lies an extra arm which consists of 3-21 bases. Depending upon the length, extra arms are of two types, small extra arm with 3-5 bases and other a large arm having 13-21 bases.
- T-arm or TψC arm:
- It has a modified base pseudouridine ψ. It has a five base pair stem with a loop.
There are about 50 different types of modified bases in different tRNAs, but four bases are more common. One is ribothymidine which contains thymine which is not found in RNA. Other modified bases are pseudouridine ψ, dihyrouridine, and inosine.
Hair Pin Structure of tRNA:
Some RNAs are Enzymes:
Recently it has been discovered that some RNAs play the role of enzymes, they are called ribozymes. Like typical enzymes, a ribozyme has an active site, a binding site for substrate and a binding site for a co-factor. Ribozymes are mainly involved in the splicing of introns present on RNA molecules.
Small Nuclear RNAs:
RNA polymerase enzyme transcribes several small RNAs in the nucleus of eukaryotes. These are called small nuclear RNAs (snRNA). These form complex with specific proteins and are called small nuclear ribonuclear proteins (snRNP) also known as snurps. The proteins associated with snRNAs are called S proteins.
These sn RNAs are rich in uracil and are of several types i.e., U1, U2, U4, U5, and U6. Each of these RNAs is between 200-300 nucleotides long. They are involved in the splicing of group II introns. They form a complex with an intron. This complex is called spliceosome which is involved in splicing of the intron.
These snRNP molecules contain small RNA sequences which are complementary to the introns of mRNA and form RNA-RNA base pairs at 5′ and 3′ splice sites where actual splice reaction occurs.
Small Nucleolar RNA (sno RNA):
This snall RNA is required for the processing of eukaryotic rRNA molecules. The snoRNAs are associated with proteins. These snoRNAs are present in the nucleolus where the processing of rRNA takes place. Ribosome is also assembled in the nucleolus.
Many small RNAs like micro RNAs (miRNAs), small interfering RNAs (siRNAs) play their role in the silencing of genes. They act on mRNA resulting in the disruption of translation.